Share this
Using VAMS to understand SARS-CoV-2, HCoV immunity
by James Rudge, PhD, Technical Director, Trajan on Oct 11, 2021 9:00:00 AM
Simultaneous measurement of multi-antibody responses from dried blood samples.
 A study article published in the July 2021 issue of Frontiers in Immunology used Mitra samples collected from 54 study volunteers to measure antibody levels (IgA, IgG and IgM) raised from N and S antigens in:
A study article published in the July 2021 issue of Frontiers in Immunology used Mitra samples collected from 54 study volunteers to measure antibody levels (IgA, IgG and IgM) raised from N and S antigens in:
pre-pandemic, SARS-CoV-2 convalescence and vaccinated cohorts.
The study, entitled “Antibody Mediated Immunity to SARS-CoV-2 and Human Coronaviruses: Multiplex Beads Assay and Volumetric Absorptive Microsampling to Generate Immune Repertoire Cartography,” employed the use of high sensitivity multiplex immunoassay to understand the immunity landscape of SARS-CoV-2 infection and antibody cross reactivity to previous coronavirus infections.
The study refers to other seasonal human coronaviruses that aren't SARS-CoV-2 as HCoVs. The study authors visually mapped significant differences in antibody levels between the three cohorts to also demonstrate the impact of vaccination in controlling the Covid-19 Pandemic and how prior exposure to previous human coronaviruses (HCoVs) impacts antibody response.
Using Existing Technology to Remotely Monitor Infectious Disease Serology
The authors of the current study paper reviewed here, M. Zand et al, had previously developed a similar technology (mPlex-Flu assay) using the Luminex Magiplex platform for simultaneously measuring IgG raised antibodies against >30 strains of influenza. A key outcome from the mPlex Flu study, as outlined in their paper published in the Journal of Translational Science, was the successful cross validation of dried capillary blood, collected remotely on Mitra® devices with VAMS® technology and compared to wet venous serum collected in clinic.
To correlate blood values to serum, the group measured hemoglobin from subaliquots from the Mitra extracts and developed an algorithm to compensate for the negative biases observed when comparing dried blood to wet serum. Moreover, the group also demonstrated high sample stability on Mitra devices, thus leading Dr. Zand and colleagues to conclude that Mitra could be used to reliably collect samples for large-scale population studies of antibody-mediated influenza immunity. They also noted that this approach was not pathogen specific but could be used to estimate “antibody mediated immunity after vaccination for any other viral pathogens.”
Are there differences in the coronavirus antibody landscape between pre-Covid, post-Covid-19, and vaccinated cohorts?
Based on the mPlex-Flu study, M. Zand et al developed a novel bead mix to measure anti-spike and anti-nucleocapsid for IgG, IgA and IgM raised against SARS-VoV-1, SARS-VoV-2, MERS and four seasonal coronavirus strains (OC43, HKU1, NL63 and 229E). Their new assay, mPlex-CoV, was highly specific and sensitive, with an assay range of 4 logs and very low inter and intra %CV. Moreover, the group utilized HCT compensation they had developed for mPlex-Flu to obtain adjusted serum values from dried Mitra blood samples. Using Mitra devices with VAMS tips, they collected samples from three cohorts (pre-Covid [n= 21], Covid-19 convalescence [n=19], and fully vaccinated [n=14]). The researchers then analyzed the data to generate immune repertoire cartography to observe differences between the three cohorts.
Key Findings & Highlights from the HCoV Study Using VAMS with the mPlex-Cov Assay
- To act as a control, using the pre-Covid cohort, the group detected IgG antibodies for both α and β human coronaviruses, for both spike and nucleocapsid. This aligned with what had been previously reported in the literature. Also, they did not detect significant antibody levels for SARS-CoV-2 (both S and N proteins). Furthermore, there was no evidence of back cross reactivity to human OC43 and NL63 coronavirus strains. Considering the homologies between the three virus types, they commented that this was a surprising observation.
- In the post-Covid / convalescent cohort, the group observed elevated levels of all three antibody classes raised against N and S proteins, not only for SARS-CoV-2, but also for the previous SARS-CoV-1 virus (responsible for the 2002–2004 SARS outbreak in southeast Asia). In this case, the researchers hypothesized that sequence homology may have played a part. Interestingly, they also observed elevated IgG, IgM and IgA antibodies against the S protein for two of the seasonal strains (OC43 and HKU1). However, for the N protein, elevated antibody levels were only observed for the two SARS-CoV strains.
- The vaccinated cohort showed statistically significant increases in SARS-CoV-2 S protein antibody levels for all three antibodies when compared to the control cohort. Interestingly, these levels were significantly higher than levels seen in the post-Covid-19 cohort. It was postulated that this was due to the effect of receiving two Covid-19 injections temporally spaced apart, where one injection acted as an immune priming event and the other as an immunity boosting event. However, the post-Covid-19 cohort would have only experienced a priming event – a result of their initial Covid-19 infection, hence their lower antibody levels. It was thus hypothesized that these antibody levels might increase if those in the post-Covid-19 (convalescence) cohort were subsequently vaccinated.
Multi-dimensional Scaling Used in the HCoV Study
To help understand the interplay between the antibodies raised against the various viral proteins in each of the three cohorts, the research group applied metric multi-dimensional scaling (MDS) using the Pearson Correlation distance; measuring IgG levels for both N and S within each subject. For example, Figure 1 shows that a good separation is seen for the S but not the N proteins. It also shows significant differences between immunity response from humoral infection versus vaccination (certainly for the S protein).
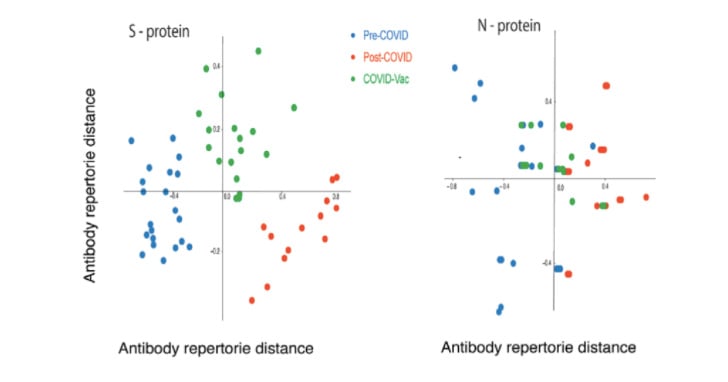 Figure 1 - MDS analysis of IgG repertoire distance from all three cohorts and between S and N proteins (Image Credit: M. Zand et al, CTSI, University of Rochester Medical Center)
Figure 1 - MDS analysis of IgG repertoire distance from all three cohorts and between S and N proteins (Image Credit: M. Zand et al, CTSI, University of Rochester Medical Center)
HCoV Study Authors’ Conclusions
The study authors concluded that Mitra devices with VAMS are an excellent approach for collecting samples remotely when risk of person-to-person infection is high. They found that combining VAMS and the mPlex-CoV assay allows for both full remote sampling as well as highly accurate lab analyses during clinical studies. Moreover, the authors commented that the combined approach also reduces the cost burden to enable future population studies. Finally, the study showed that the ability to simultaneously measure three classes of antibody raised against a range of the six most common HCoV strains will act to further our understanding of the humoral immune response to Covid-19 infection from SARS-CoV-2.
Final Thoughts
The research study reviewed here shows the excellent utility combining two cutting-edge technologies, Mitra with VAMS and mPlex-Cov, can help us gain a deeper understanding of the effect the SARS-CoV-2 virus has on antibody responses. This research paper has demonstrated that a wealth of information can be gathered by analyzing a tiny amount of remotely collected blood on a sensitive multiplex immunoassay. This combined approach could potentially pave the way towards the future of population screening (employing larger and larger cohorts).
Finally, this approach will potentially allow us to garner a deeper understanding of vaccine efficacy and help to clarify how prior exposure to related pathogens can impact the immune response to exposure of future novel viruses. This is particularly relevant now as the world becomes more globalized and as we encroach further into animal habitats, increasing the likelihood of animal-human crossover infection (zoonotic transmission) with novel pathogens.
Indeed, we have seen from Covid-19 how zoonotic transmission can wreak havoc on populations. Technologies such as Mitra and VAMS, coupled with mPlex-Cov, will help enable more remote monitoring of emerging infections and, when needed, full population serological screening research and related initiatives.
This study paper was summarized for our readers by James Rudge, PhD, Neoteryx Technical Director, as curated content. To learn more about the study conducted by researchers in the Department of Medicine, Division of Nephrology, and in the Clinical and Translational Science Institute at the University of Rochester Medical Center in Rochester, NY, please see the original article published in Frontiers in Immunology.

Share this
- Microsampling (206)
- Research, Remote Research (119)
- Venipuncture Alternative (105)
- Clinical Trials, Clinical Research (83)
- Mitra® Device (73)
- Therapeutic Drug Monitoring, TDM (51)
- Dried Blood Spot, DBS (39)
- Biomonitoring, Health, Wellness (30)
- Infectious Disease, Vaccines, COVID-19 (24)
- Blood Microsampling, Serology (23)
- Omics, Multi-Omics (21)
- Decentralized Clinical Trial (DCT) (20)
- Specimen Collection (18)
- Toxicology, Doping, Drug/Alcohol Monitoring, PEth (17)
- Skin Microsampling, Microbiopsy (14)
- hemaPEN® Device (13)
- Preclinical Research, Animal Studies (12)
- Pharmaceuticals, Drug Development (9)
- Harpera Device (7)
- Industry News, Microsampling News (5)
- Antibodies, MAbs (3)
- Company Press Release, Product Press Release (3)
- Environmental Toxins, Exposures (1)
- July 2025 (1)
- May 2025 (1)
- April 2025 (2)
- December 2024 (2)
- November 2024 (1)
- October 2024 (3)
- September 2024 (1)
- June 2024 (1)
- May 2024 (1)
- April 2024 (4)
- March 2024 (1)
- February 2024 (2)
- January 2024 (4)
- December 2023 (3)
- November 2023 (3)
- October 2023 (3)
- September 2023 (3)
- July 2023 (3)
- June 2023 (2)
- April 2023 (2)
- March 2023 (2)
- February 2023 (2)
- January 2023 (3)
- December 2022 (2)
- November 2022 (3)
- October 2022 (4)
- September 2022 (3)
- August 2022 (5)
- July 2022 (2)
- June 2022 (2)
- May 2022 (4)
- April 2022 (3)
- March 2022 (3)
- February 2022 (4)
- January 2022 (5)
- December 2021 (3)
- November 2021 (5)
- October 2021 (3)
- September 2021 (3)
- August 2021 (4)
- July 2021 (4)
- June 2021 (4)
- May 2021 (4)
- April 2021 (3)
- March 2021 (5)
- February 2021 (4)
- January 2021 (4)
- December 2020 (3)
- November 2020 (5)
- October 2020 (4)
- September 2020 (3)
- August 2020 (3)
- July 2020 (6)
- June 2020 (4)
- May 2020 (4)
- April 2020 (3)
- March 2020 (6)
- February 2020 (3)
- January 2020 (4)
- December 2019 (5)
- November 2019 (4)
- October 2019 (2)
- September 2019 (4)
- August 2019 (4)
- July 2019 (3)
- June 2019 (7)
- May 2019 (6)
- April 2019 (5)
- March 2019 (6)
- February 2019 (5)
- January 2019 (8)
- December 2018 (3)
- November 2018 (4)
- October 2018 (7)
- September 2018 (6)
- August 2018 (5)
- July 2018 (8)
- June 2018 (6)
- May 2018 (5)
- April 2018 (6)
- March 2018 (4)
- February 2018 (6)
- January 2018 (4)
- December 2017 (2)
- November 2017 (3)
- October 2017 (2)
- September 2017 (4)
- August 2017 (2)
- July 2017 (4)
- June 2017 (5)
- May 2017 (6)
- April 2017 (6)
- March 2017 (5)
- February 2017 (4)
- January 2017 (1)
- July 2016 (3)
- May 2016 (1)
- April 2016 (2)


No Comments Yet
Let us know what you think