Share this
measuring lipidomic profiles before and after exercise
by James Rudge, PhD, Technical Director, Trajan on Jul 24, 2023 9:00:00 AM
In this blog we review a follow-up study on lipidomics that was conducted by Marianne Fillet et al at the University of Liège in Belgium. In a previous blog we explored an initial lipidomics research study by Dr. Fillet et al that was published in the May 2021 edition of Frontiers in Molecular Biosciences.
 The article on that initial study discussed a proof-of-concept fluxomic study, using blood microsampling for omics studies collected with a hemaPEN® device.
The article on that initial study discussed a proof-of-concept fluxomic study, using blood microsampling for omics studies collected with a hemaPEN® device.
In the previous study, samples from 20 healthy volunteers were collected at the following time points: before, during and after running 3 lengths (400, 800 and 1600 meters) and in between short rest periods.
The biomarkers analyzed were all small metabolites, such as amino acids and organic acids. The research group found that the metabolites creatinine, choline, and taurine were significantly increased during exercise.
Choosing a Microsampling Device to Measure Lipidomic Changes
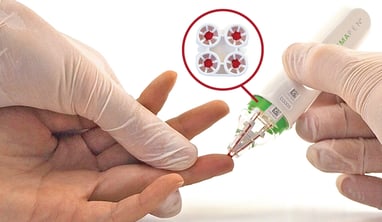 The group chose to use the hemaPEN microsampling device for their studies because it collects exactly 4 x 2.74 µL replicate samples onto dried blood spot (DBS) filter paper.
The group chose to use the hemaPEN microsampling device for their studies because it collects exactly 4 x 2.74 µL replicate samples onto dried blood spot (DBS) filter paper.
In their previous study paper, the group concluded that the hemaPEN device “offers many advantages compared to classical blood sampling.”
During their first metabolomics study using the hemaPEN, the group analyzed just one dried blood spot sample from each hemaPEN device, and they stored the other samples at –20 °C. This allowed the group to conduct the follow-up study reviewed in this blog, which was published in the Journal of Pharmaceutical and Biomedical Analysis.
The follow-up study is entitled “A targeted UHPLC-MS/MS method to monitor lipidomic changes during a physical effort: Optimization and application to blood microsamples from athletes.” In this paper, the group reported that by using the stored samples, they were able to successfully measure changes in lipidomic profiles (for 5 out of 11 targeted analytes) during the exercise program.
Why analyze the lipidome?
The lipidome is part of the wider metabolome, which consists of thousands of lipid molecules from around 15 chemical classes. These make up two thirds of the wider metabolome.
Dr. Fillet and colleagues believe that understanding the role of the lipidome in diseases such as cancer, helps to better understand the pathology of such diseases. They reported that lipidomics can also be used to evaluate the effect that sport has on the human body. They decided to focus their studies on lipidomic changes during exercise.
However, understanding the effect that exercise has on biomarkers is difficult, due to the competing factors that affect metabolic pathways. Some of these factors include, for example: age, gender, health, and nutritional status. During exercise, when glucose, (our primary source of energy) is depleted, the body turns to fatty acid metabolism for its energy. Once fatty acids are depleted, protein and amino acids are then consumed, which is not desired for muscle tone.
To evaluate the effect of exercise on lipid biomarker levels, the group decided to measure blood samples obtained from the previously collected hemaPEN samples they had used in their first study. The group targeted three classes of lipid biomarkers: Lysophospholipids (LPs, found in cellular membranes), sphingolipids (SGs) and fatty acids (FFAs).
Study Authors' Highlights: Results and Discussion
- Two physical extraction methods were evaluated: thermo-mixing and then thermo-mixing combined with a separate sonication step. A range of extraction solvent mixtures, were tested to find the optimum conditions, including methanol water, ethanol water, methanol TBME and ethanol TBME. The best recoveries were from MeOH:water (80:20). These were then dried down and reconstituted before analysis by LC-MS/MS.
- This resulted in CVs < 15% for most of the compounds.
- Extraction recoveries were > 50% for the majority of compounds.
- This resulted in CVs < 15% for most of the compounds.
- High ion suppression was observed for a number of the analytes. It was hypothesized that this was a result of competition in the source for analytes, which were not being targeted by the MS.
- It is well known that the biomarker lactic acid increases with exercise (as a result of aerobic respiration shifting to anaerobic respiration) and settles down at rest (after the oxygen debit is repaid). The researchers thus used this as a ‘control’ to compare and contrast the effect of the lipidomic biomarkers in the study using the volunteer blood samples.
- For the lysophosphatidylcholine (Lyso PC) biomarkers, a significant statistical decrease was seen for 14:0 Lyso PC and 18:1 Lyso PC. This would be expected, as it has been shown previously that with exercise, these biomarkers would indeed decrease. However, in this study, not all Lyso PCs showed this trend. It was thought that this was due to the apparent short duration of the exercise compared to what had been reported in previous published studies.
- A significant increase in arachidonic acid was observed and this reflects findings in the literature that fatty acid (FA) levels increase with exercise. It must be noted that in this study not all the FA which were tested had increased. It was concluded that, like the Lyso PC biomarkers, the reason not as many FAs had shown a change as assumed, was also due to the shorter duration of the exercise compared to other reported studies.
- In terms of the sphingolipids, the literature is inconclusive as to whether this specific class of biomarkers increases or decreases with exercise. However, in the reported study, sphingosine was significantly increased during exercise but then settled at final rest.
Study Authors’ Conclusions
Significant changes in 5 of the 11 analytes were seen, which was consistent with the literature. This study demonstrated the utility of employing the portable hemaPEN microsampling device for collecting blood samples as an alternative to traditional phlebotomy.
This approach also opens the possibility of untargeted lipidomics to help further understand metabolic pathways. The methods used in this study could potentially be used to screen for specific diseases from self-collected samples.
Neoteryx Comments
The work conducted by Marianne Fillet et al further illustrates the utility of capillary blood microsampling in Omics research. The advantage that a device like the hemaPEN is that it can provide 4 replicates per sampling event, some of the samples could be stored for future studies. This then allowed for multiple separate study papers to be published without the need to repeat the collection event.
This article was summarized for our readers by James Rudge, PhD, Microsampling Technical Director. This is curated content. To learn more about the important research outlined in this blog, visit the original article in the Journal of Pharmaceutical and Biomedical Analysis.
Image Credits: Trajan, Neoteryx, iStock
Share this
- Microsampling (206)
- Research, Remote Research (119)
- Venipuncture Alternative (105)
- Clinical Trials, Clinical Research (83)
- Mitra® Device (73)
- Therapeutic Drug Monitoring, TDM (51)
- Dried Blood Spot, DBS (39)
- Biomonitoring, Health, Wellness (30)
- Infectious Disease, Vaccines, COVID-19 (24)
- Blood Microsampling, Serology (23)
- Omics, Multi-Omics (21)
- Decentralized Clinical Trial (DCT) (20)
- Specimen Collection (18)
- Toxicology, Doping, Drug/Alcohol Monitoring, PEth (17)
- Skin Microsampling, Microbiopsy (14)
- hemaPEN® Device (13)
- Preclinical Research, Animal Studies (12)
- Pharmaceuticals, Drug Development (9)
- Harpera Device (7)
- Industry News, Microsampling News (5)
- Antibodies, MAbs (3)
- Company Press Release, Product Press Release (3)
- Environmental Toxins, Exposures (1)
- July 2025 (1)
- May 2025 (1)
- April 2025 (2)
- December 2024 (2)
- November 2024 (1)
- October 2024 (3)
- September 2024 (1)
- June 2024 (1)
- May 2024 (1)
- April 2024 (4)
- March 2024 (1)
- February 2024 (2)
- January 2024 (4)
- December 2023 (3)
- November 2023 (3)
- October 2023 (3)
- September 2023 (3)
- July 2023 (3)
- June 2023 (2)
- April 2023 (2)
- March 2023 (2)
- February 2023 (2)
- January 2023 (3)
- December 2022 (2)
- November 2022 (3)
- October 2022 (4)
- September 2022 (3)
- August 2022 (5)
- July 2022 (2)
- June 2022 (2)
- May 2022 (4)
- April 2022 (3)
- March 2022 (3)
- February 2022 (4)
- January 2022 (5)
- December 2021 (3)
- November 2021 (5)
- October 2021 (3)
- September 2021 (3)
- August 2021 (4)
- July 2021 (4)
- June 2021 (4)
- May 2021 (4)
- April 2021 (3)
- March 2021 (5)
- February 2021 (4)
- January 2021 (4)
- December 2020 (3)
- November 2020 (5)
- October 2020 (4)
- September 2020 (3)
- August 2020 (3)
- July 2020 (6)
- June 2020 (4)
- May 2020 (4)
- April 2020 (3)
- March 2020 (6)
- February 2020 (3)
- January 2020 (4)
- December 2019 (5)
- November 2019 (4)
- October 2019 (2)
- September 2019 (4)
- August 2019 (4)
- July 2019 (3)
- June 2019 (7)
- May 2019 (6)
- April 2019 (5)
- March 2019 (6)
- February 2019 (5)
- January 2019 (8)
- December 2018 (3)
- November 2018 (4)
- October 2018 (7)
- September 2018 (6)
- August 2018 (5)
- July 2018 (8)
- June 2018 (6)
- May 2018 (5)
- April 2018 (6)
- March 2018 (4)
- February 2018 (6)
- January 2018 (4)
- December 2017 (2)
- November 2017 (3)
- October 2017 (2)
- September 2017 (4)
- August 2017 (2)
- July 2017 (4)
- June 2017 (5)
- May 2017 (6)
- April 2017 (6)
- March 2017 (5)
- February 2017 (4)
- January 2017 (1)
- July 2016 (3)
- May 2016 (1)
- April 2016 (2)



No Comments Yet
Let us know what you think